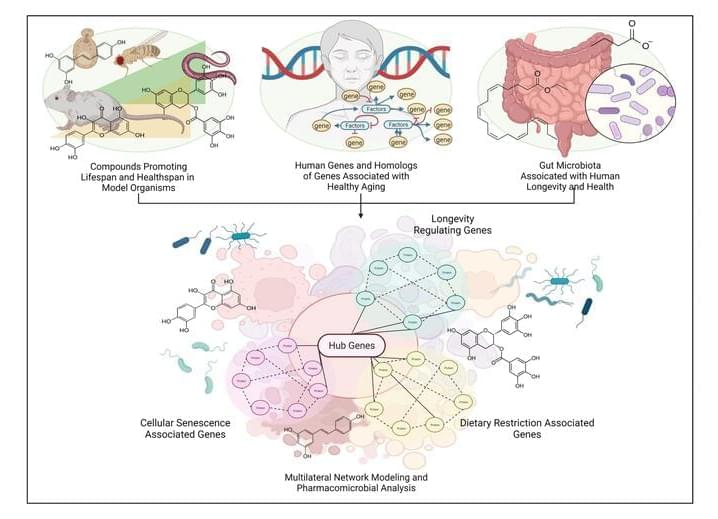
Advances in antiaging drug/lead discovery in animal models constitute a large body of literature on novel senotherapeutics and geroprotectives. However, with little direct evidence or mechanism of action in humans—these drugs are utilized as nutraceuticals or repurposed supplements without proper testing directions, appropriate biomarkers, or consistent in-vivo models. In this study, we take previously identified drug candidates that have significant evidence of prolonging lifespan and promoting healthy aging in model organisms, and simulate them in human metabolic interactome networks. Screening for drug-likeness, toxicity, and KEGG network correlation scores, we generated a library of 285 safe and bioavailable compounds. We interrogated this library to present computational modeling-derived estimations of a tripartite interaction map of animal geroprotective compounds in the human molecular interactome extracted from longevity, senescence, and dietary restriction-associated genes. Our findings reflect previous studies in aging-associated metabolic disorders, and predict 25 best-connected drug interactors including Resveratrol, EGCG, Metformin, Trichostatin A, Caffeic Acid and Quercetin as direct modulators of lifespan and healthspan-associated pathways. We further clustered these compounds and the functionally enriched subnetworks therewith to identify longevity-exclusive, senescence-exclusive, pseudo-omniregulators and omniregulators within the set of interactome hub genes. Additionally, serum markers for drug-interactions, and interactions with potentially geroprotective gut microbial species distinguish the current study and present a holistic depiction of optimum gut microbial alteration by candidate drugs. These findings provide a systems level model of animal life-extending therapeutics in human systems, and act as precursors for expediting the ongoing global effort to find effective antiaging pharmacological interventions.
Communicated by Ramaswamy H. Sarma.









Comments are closed.