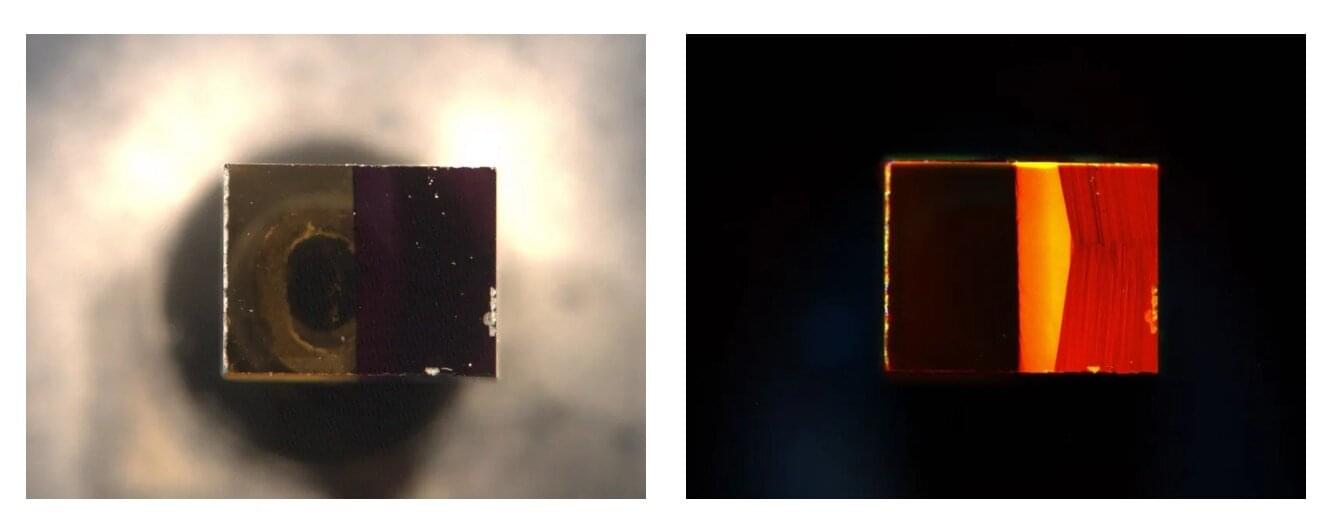
Researchers at HZDR have partnered with the Norwegian University of Science and Technology in Trondheim, and the Institute of Nuclear Physics in the Polish Academy of Sciences to develop a method that facilitates the manufacture of particularly efficient magnetic nanomaterials in a relatively simple process based on inexpensive raw materials.
Using a highly focused ion beam, they imprint magnetic nanostrips consisting of tiny, vertically aligned nanomagnets onto the materials. As the researchers have reported in the journal Advanced Functional Materials, this geometry makes the material highly sensitive to external magnetic fields and current pulses.
Nanomagnets play a key role in modern information technologies. They facilitate fast data storage, precise magnetic sensors, novel developments in spintronics, and, in the future, quantum computing. The foundations of all these applications are functional materials with particular magnetic structures that can be customized on the nanoscale and precisely controlled.