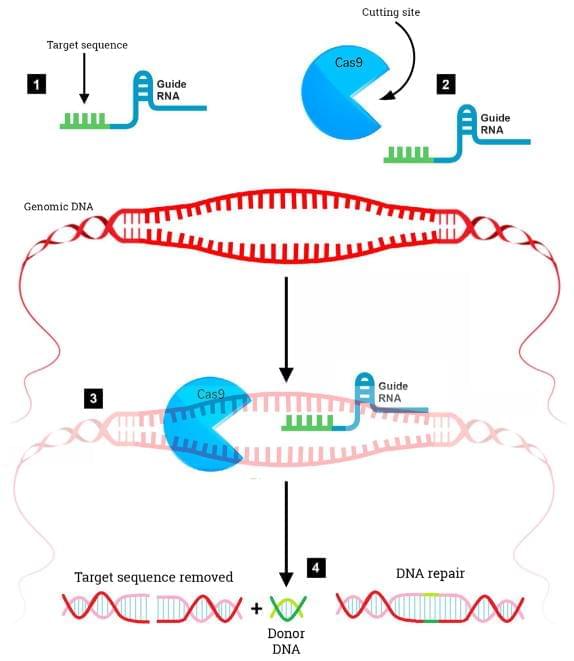
Despite a wealth of knowledge gained in the past three decades concerning the molecular underpinnings of Alzheimer’s disease (AD), progress towards obtaining effective, disease modifying therapies has proven to be challenging. In this manner, numerous clinical trials targeting the production, aggregation, and toxicity of beta-amyloid, have failed to meet efficacy standards. This puts into question the beta-amyloid hypothesis and suggests that additional treatment strategies should be explored. The recent emergence of CRISPR/Cas9 gene editing as a relatively straightforward, inexpensive, and precise system has led to an increased interest of applying this technique in AD. CRISPR/Cas9 gene editing can be used as a direct treatment approach or to help establish better animal models that more faithfully mimic human neurodegenerative diseases. In this manner, this technique has already shown promise in other neurological disorders, such as Huntington’s disease. The purpose of this review is to examine the potential utility of CRISPR/Cas9 as a treatment option for AD by targeting specific genes including those that cause early-onset AD, as well as those that are significant risk factors for late-onset AD such as the apolipoprotein E4 (APOE4) gene.
Keywords: Alzheimer’s disease, CRISPR/Cas9, Gene editing, Treatment, Huntington’s disease, iPSC neurons.
Alzheimer’s Disease (AD) is a progressive and fatal neurodegenerative disorder that primarily affects older adults and is the most common cause of dementia [1]. Currently it afflicts 5.5 million Americans and that number is expected to triple by 2050. At the present time, it is the third leading cause of death behind heart disease and cancer, with an estimated 700,000 Americans ages65 years will have AD when they die [2]. In addition, the cost of the disease is substantial with $259 billion health care dollars going to manage the disease currently, and by the middle of the century costs are predicted to soar over $1.2 trillion, which will completely bankrupt the healthcare system in the USA [3]. Worldwide, 47 million people live with dementia and that number is projected to increase to more than 131 million by 2050 with an estimated worldwide cost of US $818 billion [4].